
Research
We would like to understand how cell identity, metabolism, and responses to environmental cues are linked to control plant development. We are particularly interested in how chromatin structure contributes to transcriptional regulation of genes involved in these processes. Our favourite chromatin-modifying complex is Polycomb Repressive Complex 2 (PRC2). This is a well-conserved histone methyltrasferase responsible for methylation of lysine 27 of histone 3 (H3K27me), an epigenetic mark associated with chromatin compaction and gene repression. PRC2 is best known for its important role in maintaining cell fate and controlling developmental transitions in multicellular species. Interestingly, it is well conserved in most eukaryotic supergroups, raising interesting questions about its origin and evolution.
PRC2-mediated regulation of seedling establishment and photoautotrophic growth
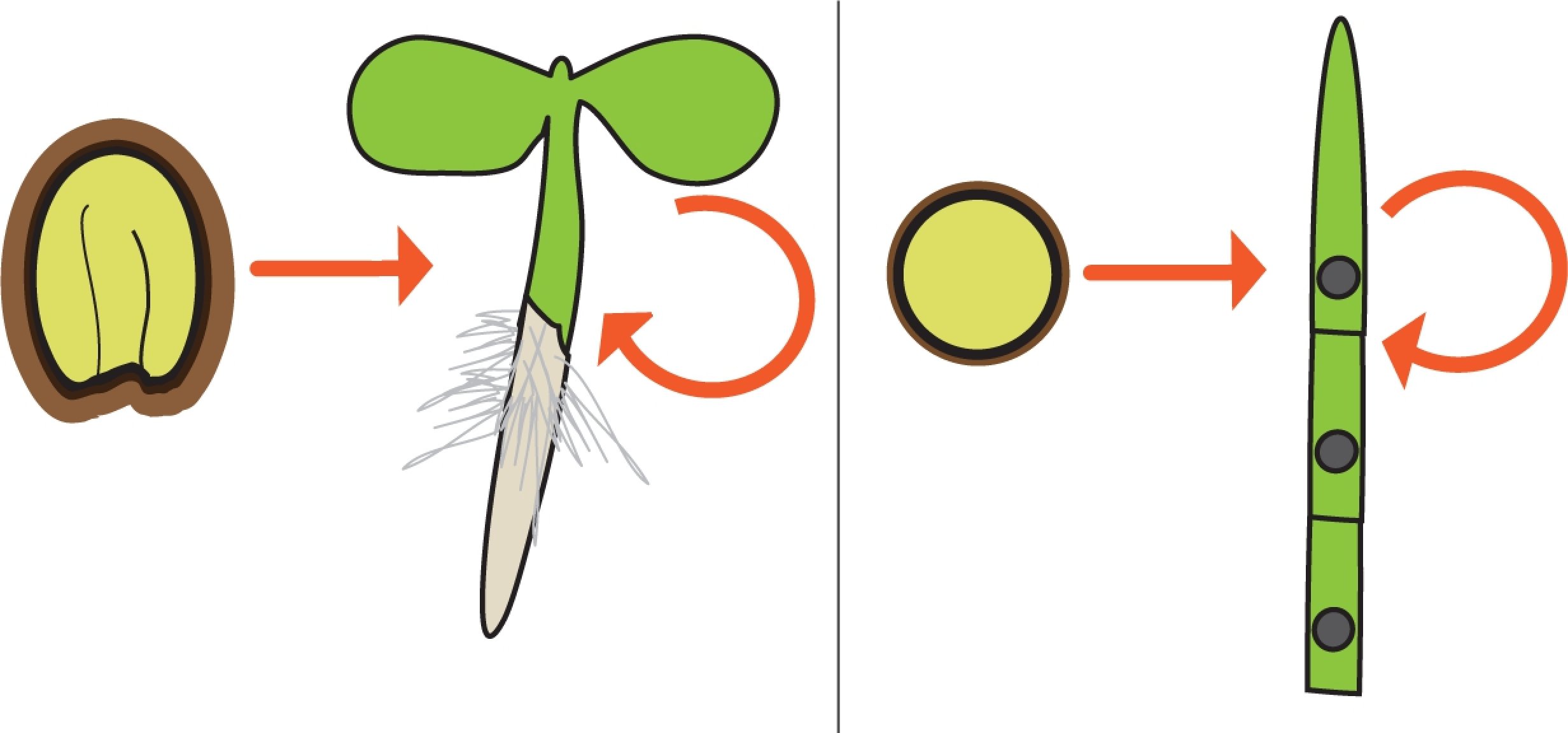 |
Seed or spore germination in land plants involves extensive reprogramming of development and metabolism, which is essential for the establishment of photoautotrophic growth. Our goal is to decipher (1) the role of PRC2 in this developmental and metabolic transition and (2) its contribution to plant acclimation to environmental conditions. We use the flowering plant Arabidopsis thaliana and the early land plant Physcomitrium patens as models to determine the extent of conservation of these regulatory mechanisms during land plant evolution. |
Origin and evolution of PRC2
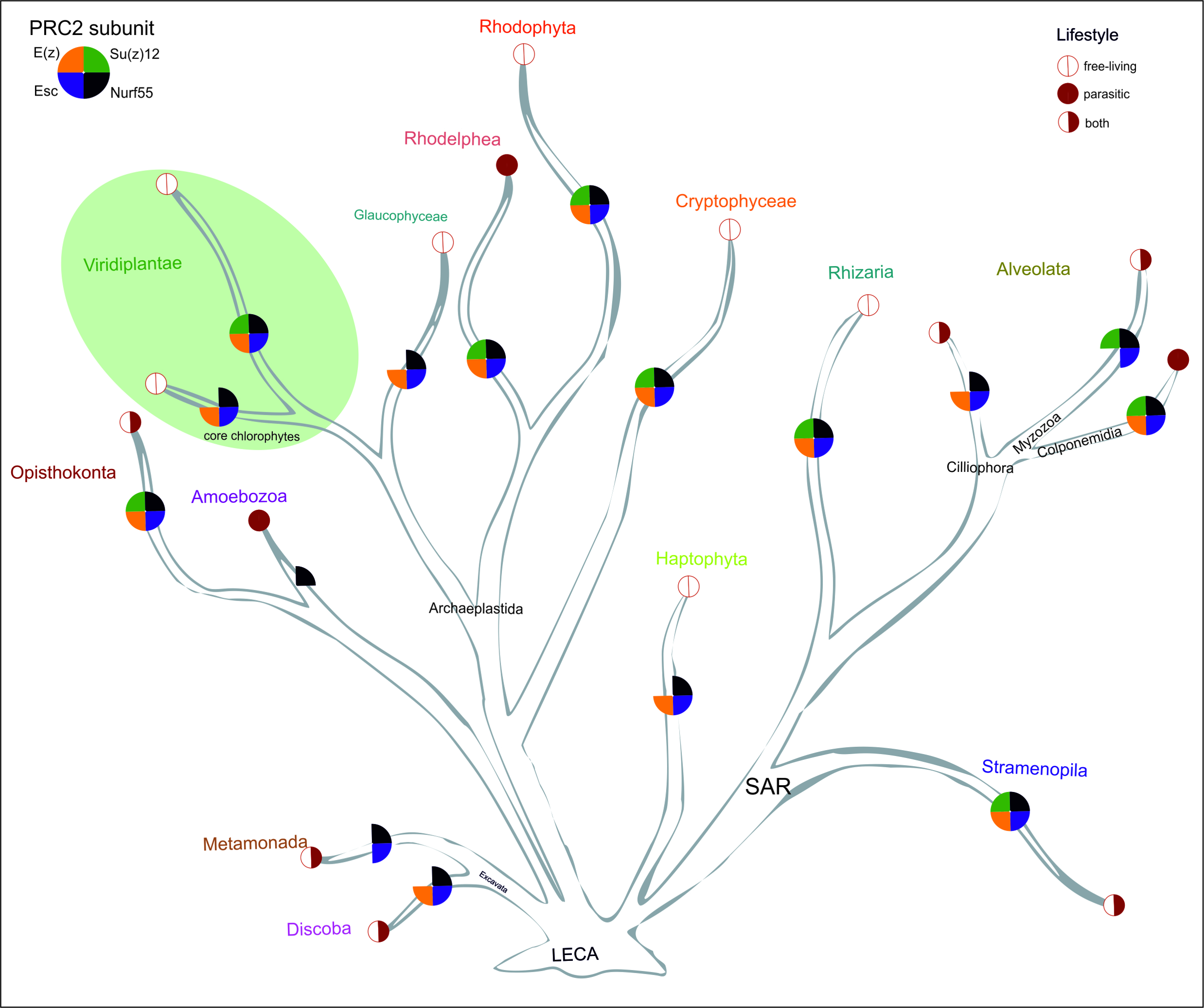 |
PRC2 is well conserved in the evolution of eukaryotes. In our work, we address the question of how the composition and function of PRC2 evolved in the green lineage and also in other eukaryotic supergroups. Recently, we developed an automated computational pipeline "PcG-finder" to identify PRC2 protein components and reconstruct the phylogeny of PRC2 core subunits in all eukaryotic lineages (Sharaf et al. 2022). This opened up intriguing new questions regarding the evolution of the protein complex composition and function that we are now addressing, mainly using the green lineage as an "experimental playground". |
|
| Image modified from Sharaf et al. 2022 PubMed |